ALLCools: ALL methyl-Cytosine tools
Contents
ALLCools: ALL methyl-Cytosine tools¶
ALLCools is a software package for single-cell DNA methylome data analysis. It contains functions for cellular and genomic analysis.
Documentation Organization¶
For installation, please read GET STARTED here;
The ALLCools workflow encompasses the following two sections:
CELLULAR ANALYSIS: This section goes over basic clustering steps, differential methylated genes (DMG) analysis, data integration, and doublets identification;
GENOMIC ANALYSIS: This section covers differential methylated region (DMR) calling, genome annotation, DNA motif analysis, correlation analysis, and enhancer prediction;
To generate analysis files from your own data, please read the COMMAND LINE TOOLS here;
For references to function used in this package, please read the API here.
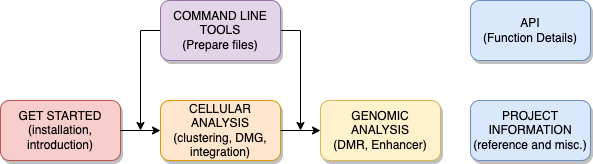
Fig. 1 ALLCools documentation organization.¶
Authors¶
Hanqing Liu, developer, initial conception
Jingtian Zhou, developer, 5kb clustering algorithms
Wei Tian
Jiaying Xu
Support¶

Fig. 2 Click on this to create a page specific issue.¶
The source code is on github;
For releases and changelog, please check out the github releases page;
For bugs and feature requests, please use the issue tracker.
For page-specific issues, please use the “open issue” button on the top-right toggle.
Citing ALLCools¶
If ALLCools has been significant in your research, and you would like to acknowledge the project in your academic publication, we suggest citing our paper [Liu et al., 2021]. For any specific methods and algorithms, also consider citing the original author’s paper included in the Citation and Reference page here.
