Preprocess SMART-seq
Contents
Preprocess SMART-seq¶
Load data¶
Gene raw counts matrix from SMART-seq
adata = anndata.read_h5ad('../input/SMART.Neuron.h5ad')
adata
AnnData object with n_obs × n_vars = 4051 × 24446
obs: 'MajorType', 'SubType'
Preprocessing¶
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=3)
adata
AnnData object with n_obs × n_vars = 4051 × 20152
obs: 'MajorType', 'SubType', 'n_genes'
var: 'n_cells'
# basic steps of the scanpy clustering process
adata.layers['raw'] = adata.X.copy()
sc.pp.normalize_per_cell(adata)
sc.pp.log1p(adata)
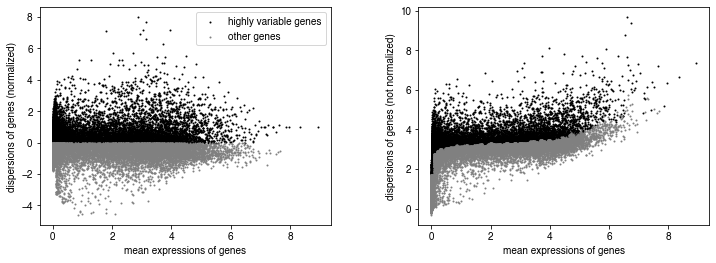
sc.pp.highly_variable_genes(adata, n_bins=100, n_top_genes=10000)
sc.pl.highly_variable_genes(adata)
Save AnnData¶
# this is the total adata without downsample
adata.write_h5ad(f'SMART.TotalAdata.norm_log1p.h5ad')
adata
AnnData object with n_obs × n_vars = 4051 × 20152
obs: 'MajorType', 'SubType', 'n_genes', 'n_counts'
var: 'n_cells', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'log1p', 'hvg'
layers: 'raw'
