Prepare Cluster-by-Gene mC Profile
Contents
Prepare Cluster-by-Gene mC Profile¶
This notebook sum cell feature counts in a cell MCDS by cell clusters. The cluster MCDS can be transform and saved as RegionDS
import pandas as pd
from ALLCools.mcds import MCDS
from ALLCools.clustering import cluster_enriched_features
Merge Cluster¶
mc_cluster = pd.read_csv(
'../../data/HIPBulk/DesideBulkLevel/snmC.anno.csv',
header=None,
index_col=0,
squeeze=True)
m3c_cluster = pd.read_csv(
'../../data/HIPBulk/DesideBulkLevel/snm3C.anno.csv',
header=None,
index_col=0,
squeeze=True)
mc_cluster = 'snmC_' + mc_cluster
m3c_cluster = 'snm3C_' + m3c_cluster
cell_cluster = pd.concat([mc_cluster, m3c_cluster])
cell_cluster.index.name = 'cell'
cell_cluster.name = 'sample'
mcds = MCDS.open('../../data/Brain/snm*C-seq*/*.mcds')
Open MCDS with netcdf4 engine.
mcds.coords['sample'] = cell_cluster
cluster_mcds = mcds.merge_cluster(cluster_col='sample').to_region_ds('geneslop2k')
cluster_mcds
<xarray.RegionDS>
Dimensions: (mc_type: 2, count_type: 2, geneslop2k: 55487, sample: 20, chrom100k: 27269)
Coordinates: (12/14)
* mc_type (mc_type) object 'CGN' 'CHN'
* count_type (count_type) object 'mc' 'cov'
strand_type <U4 'both'
* geneslop2k (geneslop2k) object 'ENSMUSG00000102693.1' ... 'ENSM...
geneslop2k_chrom (geneslop2k) object 'chr1' 'chr1' ... 'chrM' 'chrM'
geneslop2k_start (geneslop2k) int64 3071252 3100015 ... 13288 13355
... ...
sample_CGN (sample) float64 0.7203 0.7387 0.7422 ... 0.7434 0.7196
sample_CHN (sample) float64 0.007835 0.02362 ... 0.01121 0.005743
* chrom100k (chrom100k) int64 0 1 2 3 4 ... 27265 27266 27267 27268
chrom100k_chrom (chrom100k) object 'chr1' 'chr1' ... 'chrY' 'chrM'
chrom100k_bin_start (chrom100k) int64 0 100000 200000 ... 91700000 0
chrom100k_bin_end (chrom100k) int64 100000 200000 ... 91744698 16299
Data variables:
geneslop2k_da (sample, geneslop2k, mc_type, count_type) float64 1....
chrom100k_da (sample, chrom100k, mc_type, count_type) float64 0.0...
geneslop2k_da_frac (sample, geneslop2k, mc_type) float64 1.252 ... 4.546
chrom100k_da_frac (sample, chrom100k, mc_type) float64 1.0 1.0 ... 5.264
Attributes:
region_dim: geneslop2kcluster_mcds.to_zarr('test_HIP_Cluster')
<xarray.backends.zarr.ZarrStore at 0x7fb99abac040>
Cluster Enrichment¶
mcds.add_mc_frac(var_dim='geneslop2k')
adata = mcds.get_adata(mc_type='CHN',
var_dim='geneslop2k',
da_suffix='frac',
obs_dim='cell',
select_hvf=False)
/home/hanliu/miniconda3/envs/allcools_new/lib/python3.8/site-packages/dask/core.py:119: RuntimeWarning: invalid value encountered in true_divide
return func(*(_execute_task(a, cache) for a in args))
/home/hanliu/miniconda3/envs/allcools_new/lib/python3.8/site-packages/dask/core.py:119: RuntimeWarning: divide by zero encountered in true_divide
return func(*(_execute_task(a, cache) for a in args))
/home/hanliu/miniconda3/envs/allcools_new/lib/python3.8/site-packages/dask/array/numpy_compat.py:39: RuntimeWarning: invalid value encountered in true_divide
x = np.divide(x1, x2, out)
adata.obs['cluster'] = adata.obs['sample'].str.split('_').str[1]
cell_ids = adata.obs.groupby('cluster').apply(lambda i: i if i.shape[
0] < 1500 else i.sample(1500)).index.get_level_values(1)
adata = adata[cell_ids, :]
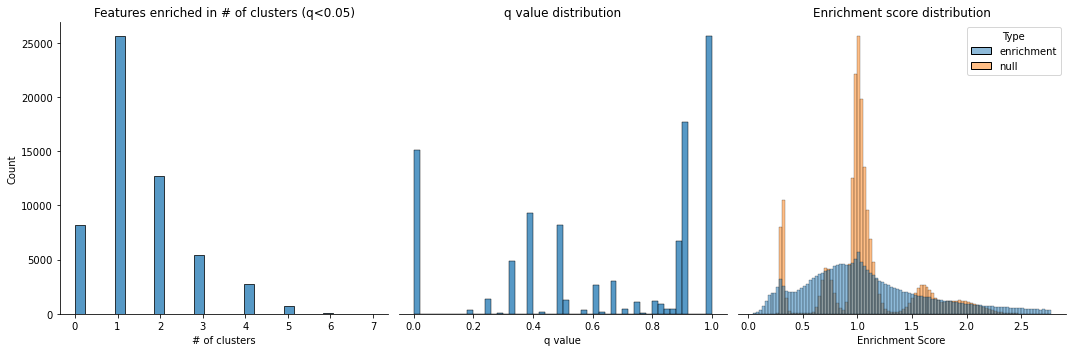
cluster_enriched_features(adata,
cluster_col='cluster',
top_n=200,
alpha=0.05,
stat_plot=True,
method='mc')
Found 10 clusters to compute feature enrichment score
Computing enrichment score
Computing enrichment score FDR-corrected P values
findfont: Font family ['sans-serif'] not found. Falling back to DejaVu Sans.
findfont: Generic family 'sans-serif' not found because none of the following families were found: Helvetica
findfont: Font family ['sans-serif'] not found. Falling back to DejaVu Sans.
findfont: Generic family 'sans-serif' not found because none of the following families were found: Helvetica
Trying to set attribute `._uns` of view, copying.
Selected 1653 unique features
qvals = pd.DataFrame(
adata.uns['cluster_feature_enrichment']['qvals'],
index=adata.var_names,
columns=adata.uns['cluster_feature_enrichment']
['cluster_order'])
qvals.to_hdf('mCH.cluster_enrichment_qvals.hdf', key='data')
