Calculate Cell Doublet Score
Contents
Calculate Cell Doublet Score¶
Content¶
Here we assign a doublet score to each cell. The MethylScrblet method we used here is adapted from the Scrublet package [Wolock et al., 2019]. High doublet score indicates that the cell is likely merged from different cell types.
Prerequisite¶
Before this step, run the Basic Clustering Walk-through to get the cell-by-feature AnnData file.
Input¶
MCDS Files (contains chrom100K dim)
Cell Metadata
Output AnnData file from Basic Clustering Walk-through.
Output¶
Cell doublet score
Import¶
import pandas as pd
import matplotlib.pyplot as plt
import anndata
from ALLCools.clustering.doublets import MethylScrublet
from ALLCools.plot import *
from ALLCools.mcds import MCDS
Parameters¶
# change this to the paths to your MCDS files
mcds_paths = '../../data/Brain/snm*C-seq*/*.mcds'
adata_path = '../step_by_step/100kb/adata.with_coords.h5ad'
cef_path = '../step_by_step/100kb/L1_enriched_features.mCH.txt'
var_dim = 'chrom100k'
obs_dim = 'cell'
cluster_col = 'L1'
load = True
mc_type = 'CHN'
# Clustering resolution
n_neighbors = 10
expected_doublet_rate=0.06
plot_type = 'static'
Load¶
AnnData with clustering results¶
adata = anndata.read_h5ad(adata_path)
use_features = pd.read_csv(cef_path, index_col=0, header=None).index
use_features.name = var_dim
Raw mC counts¶
mcds = MCDS.open(mcds_paths, obs_dim=obs_dim, use_obs=adata.obs_names)
adata = adata[mcds.get_index(obs_dim), :].copy()
mc = mcds['chrom100k_da'].sel({
'mc_type': mc_type,
'count_type': 'mc',
use_features.name: use_features
})
cov = mcds['chrom100k_da'].sel({
'mc_type': mc_type,
'count_type': 'cov',
use_features.name: use_features
})
if load and (mcds.get_index('cell').size <= 20000):
mc.load()
cov.load()
Calculate Doublet Scores¶
scrublet = MethylScrublet(sim_doublet_ratio=2.0,
n_neighbors=n_neighbors,
expected_doublet_rate=expected_doublet_rate,
stdev_doublet_rate=0.02,
metric='euclidean',
random_state=0,
n_jobs=-1)
score, judge = scrublet.fit(mc, cov, clusters=adata.obs[cluster_col])
adata.obs['doublet_score'] = score
adata.obs['is_doublet'] = judge
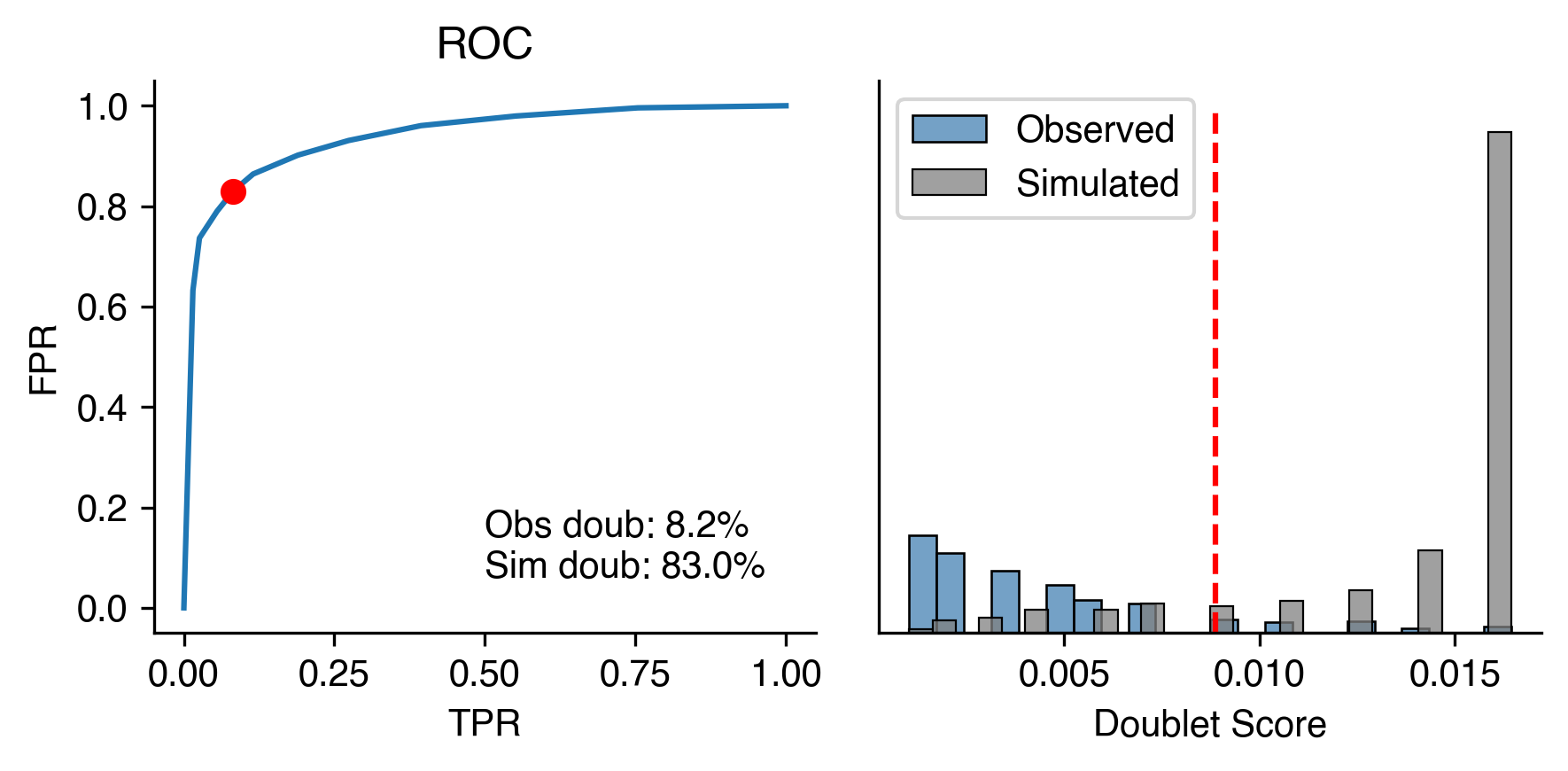
scrublet.plot()
Calculating mC frac of observations...
Simulating doublets...
Cell cluster labels are given, will sample similar number of cells from each cluster.
PCA...
Calculating doublet scores...
Automatically set threshold to 0.01
Detected doublet rate = 8.2%
Estimated detectable doublet fraction = 83.0%
Overall doublet rate:
Expected = 6.0%
Estimated = 9.8%
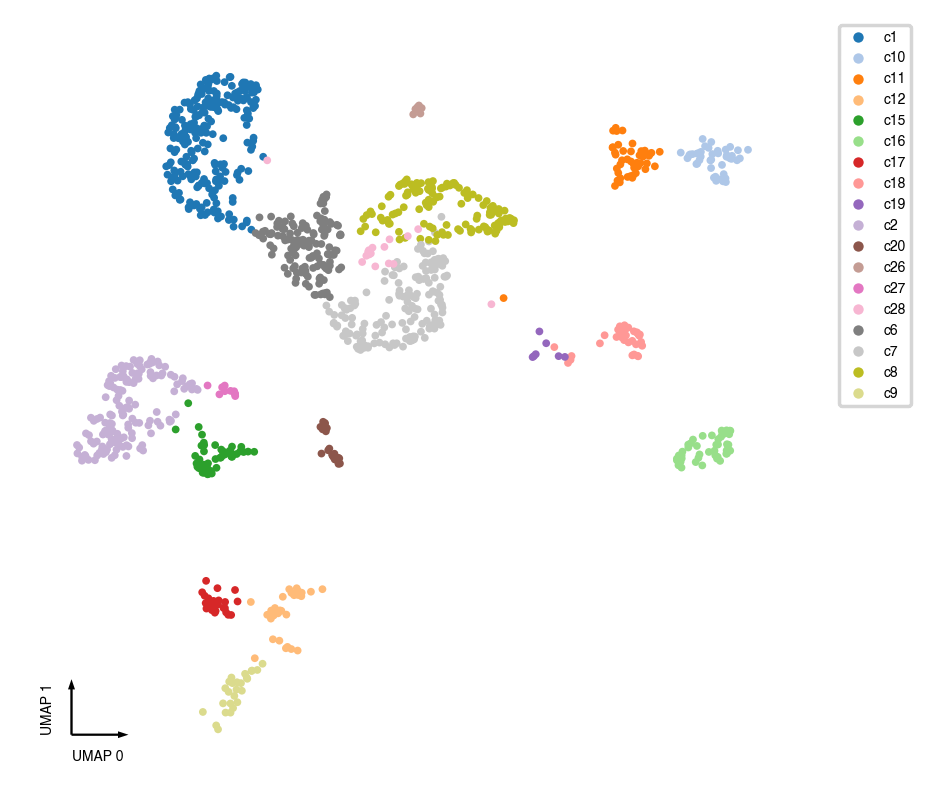
fig, ax = plt.subplots(figsize=(4, 4), dpi=250)
_ = categorical_scatter(data=adata.obs, ax=ax, coord_base='umap', hue=cluster_col, show_legend=True)
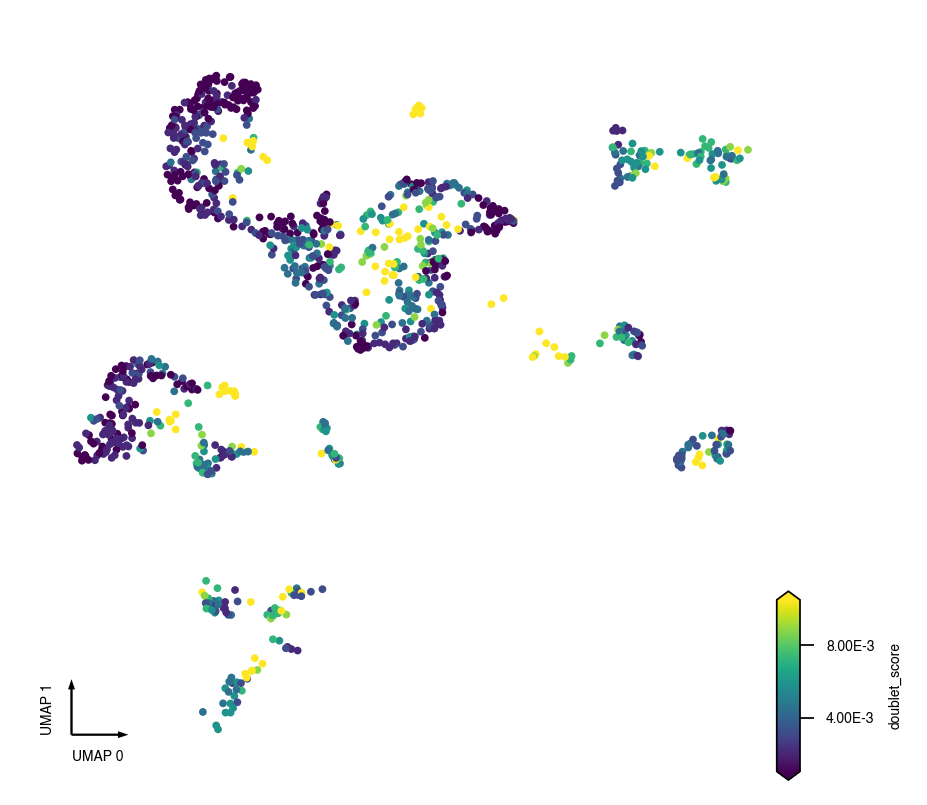
fig, ax = plt.subplots(figsize=(4, 4), dpi=250)
_ = continuous_scatter(data=adata.obs, ax=ax, coord_base='umap', hue='doublet_score')
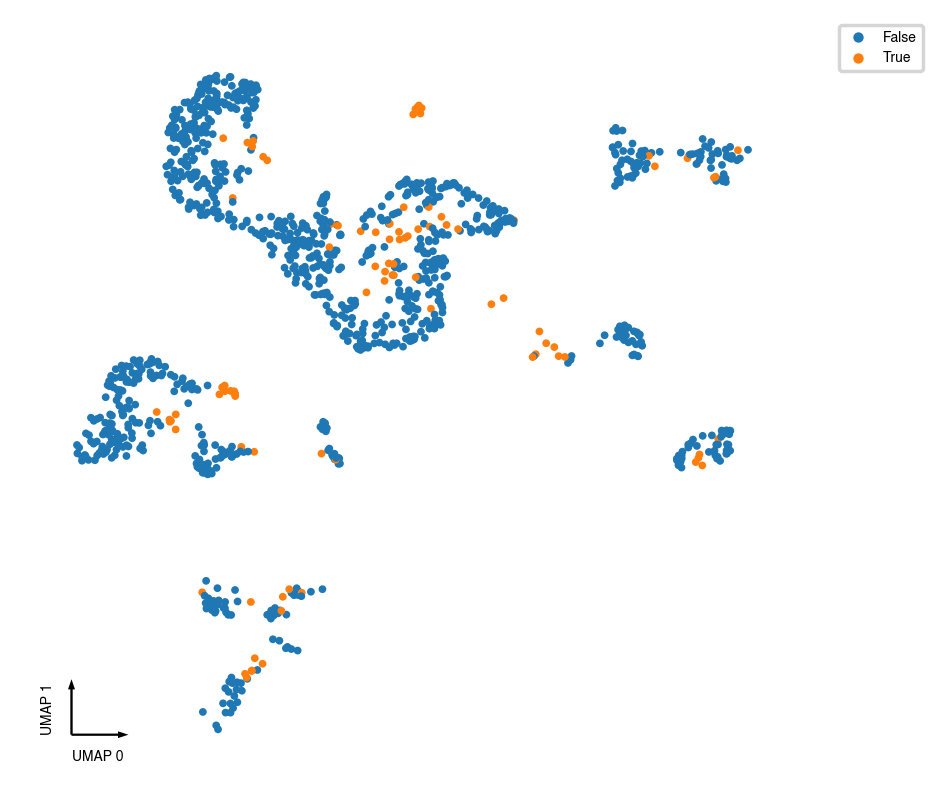
Plots¶
fig, ax = plt.subplots(figsize=(4, 4), dpi=250)
_ = categorical_scatter(data=adata.obs,
ax=ax,
coord_base='umap',
hue='CellTypeAnno',
text_anno='CellTypeAnno',
palette='tab20',
show_legend=True)

fig, ax = plt.subplots(figsize=(4, 4), dpi=250)
_ = continuous_scatter(data=adata.obs, ax=ax, coord_base='umap', hue='doublet_score')
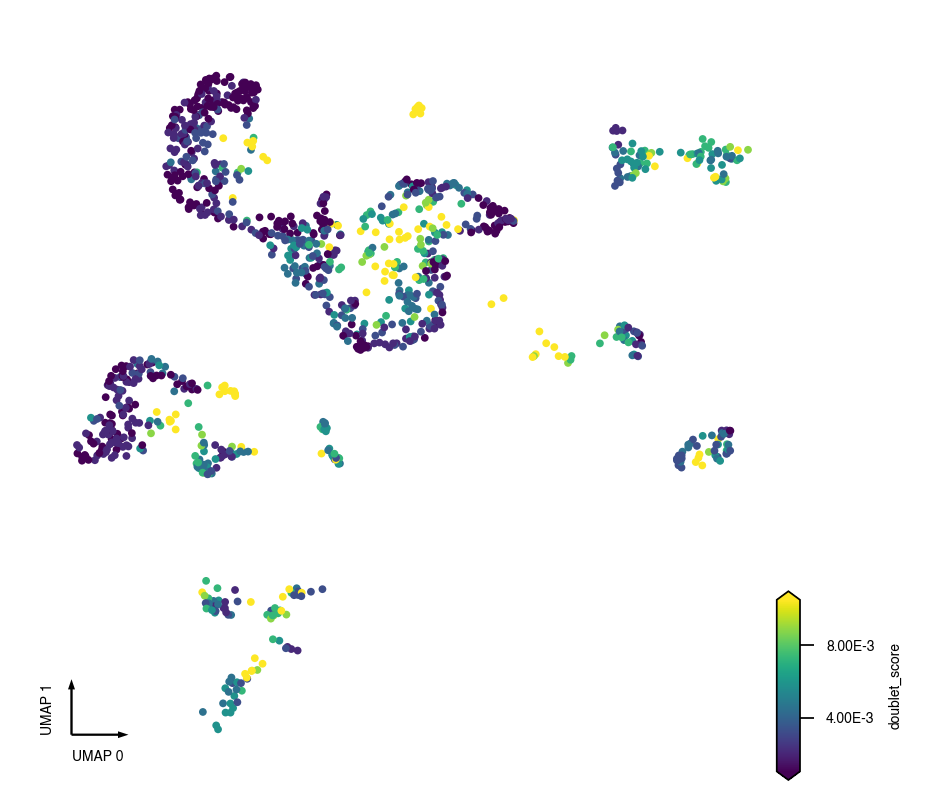
fig, ax = plt.subplots(figsize=(4, 4), dpi=250)
_ = categorical_scatter(data=adata.obs, ax=ax, coord_base='umap', hue='is_doublet', show_legend=True)