Preclustering and Cluster Enriched Features
Contents
Preclustering and Cluster Enriched Features¶
Purpose¶
The purpose of this step is to perform a simple pre-clustering using the highly variable features to get a pre-clusters labeling. We then select top enriched features for each cluster (CEF) for further analysis.
Input¶
HVF adata file.
Output¶
HVF adata file with pre-clusters and CEF annotated.
Import¶
import seaborn as sns
import anndata
import scanpy as sc
from ALLCools.clustering import cluster_enriched_features, significant_pc_test, log_scale
sns.set_context(context='notebook', font_scale=1.3)
Parameters¶
adata_path = 'mCG.HVF.h5ad'
# Cluster Enriched Features analysis
top_n=200
alpha=0.05
stat_plot=True
# you may provide a pre calculated cluster version.
# If None, will perform basic clustering using parameters below.
cluster_col = None
# These parameters only used when cluster_col is None
k=25
resolution=1
cluster_plot=True
Load Data¶
adata = anndata.read_h5ad(adata_path)
Pre-Clustering¶
If cluster label is not provided, will perform basic clustering here
if cluster_col is None:
# IMPORTANT
# put the unscaled matrix in adata.raw
adata.raw = adata
log_scale(adata)
sc.tl.pca(adata, n_comps=100)
significant_pc_test(adata, p_cutoff=0.1, update=True)
sc.pp.neighbors(adata, n_neighbors=k)
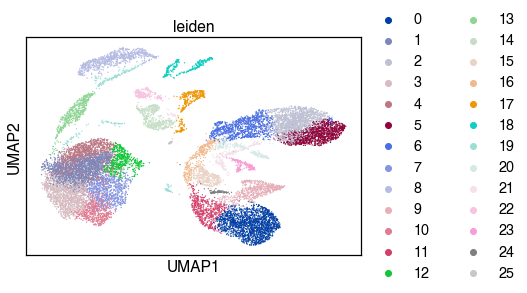
sc.tl.leiden(adata, resolution=resolution)
if cluster_plot:
sc.tl.umap(adata)
sc.pl.umap(adata, color='leiden')
# return to unscaled X, CEF need to use the unscaled matrix
adata = adata.raw.to_adata()
cluster_col = 'leiden'
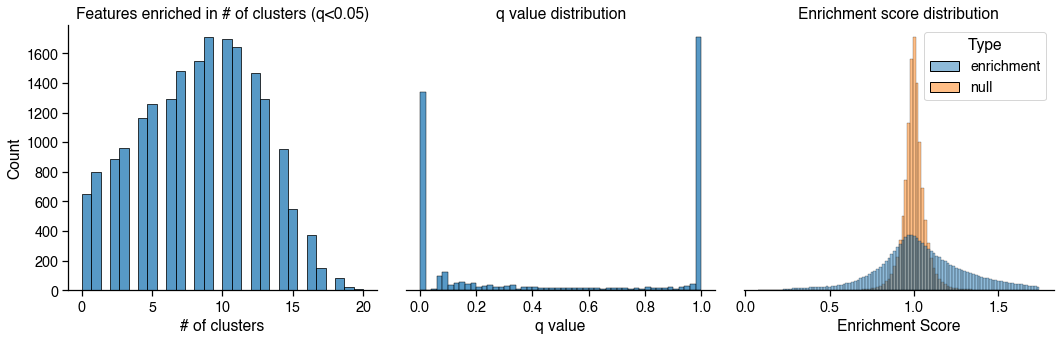
Cluster Enriched Features (CEF)¶
cluster_enriched_features(adata,
cluster_col=cluster_col,
top_n=top_n,
alpha=alpha,
stat_plot=True)
Found 26 clusters to compute feature enrichment score
Computing enrichment score
Computing enrichment score FDR-corrected P values
Selected 2379 unique features
Save AnnData¶
# save adata
adata.write_h5ad(adata_path)
adata
AnnData object with n_obs × n_vars = 16985 × 20000
obs: 'leiden'
var: 'bin_end', 'bin_start', 'chrom', 'CHN_mean', 'CHN_dispersion', 'CHN_cov', 'CHN_score', 'CHN_feature_select', 'CGN_mean', 'CGN_dispersion', 'CGN_cov', 'CGN_score', 'CGN_feature_select', 'leiden_enriched_features'
uns: 'log', 'pca', 'neighbors', 'leiden', 'umap', 'leiden_colors', 'leiden_feature_enrichment'
obsm: 'X_pca', 'X_umap'
